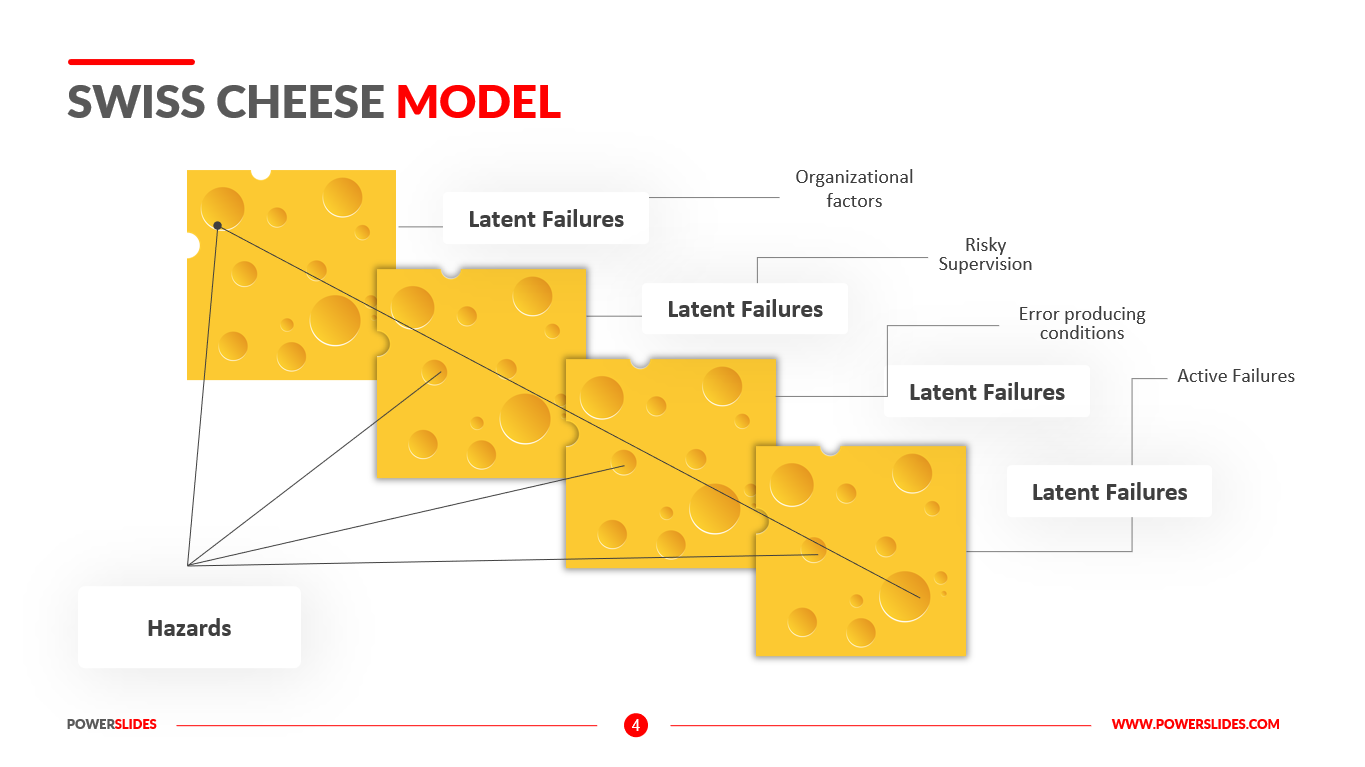
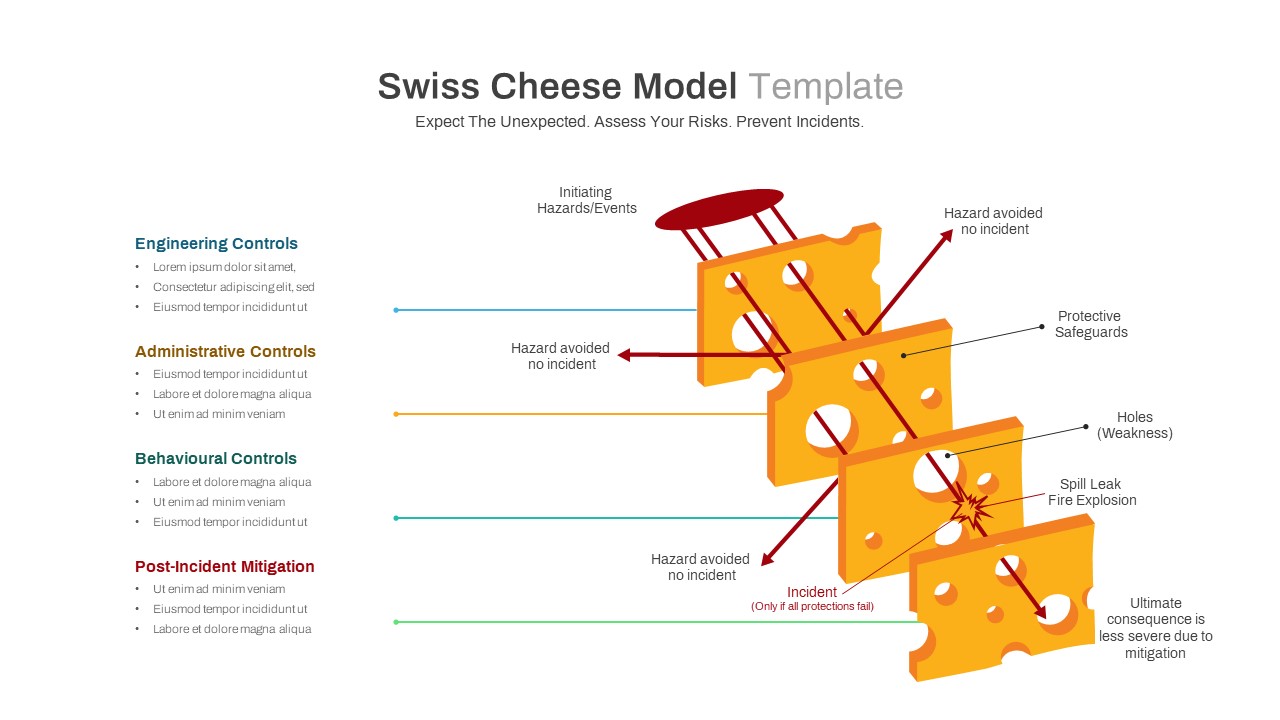
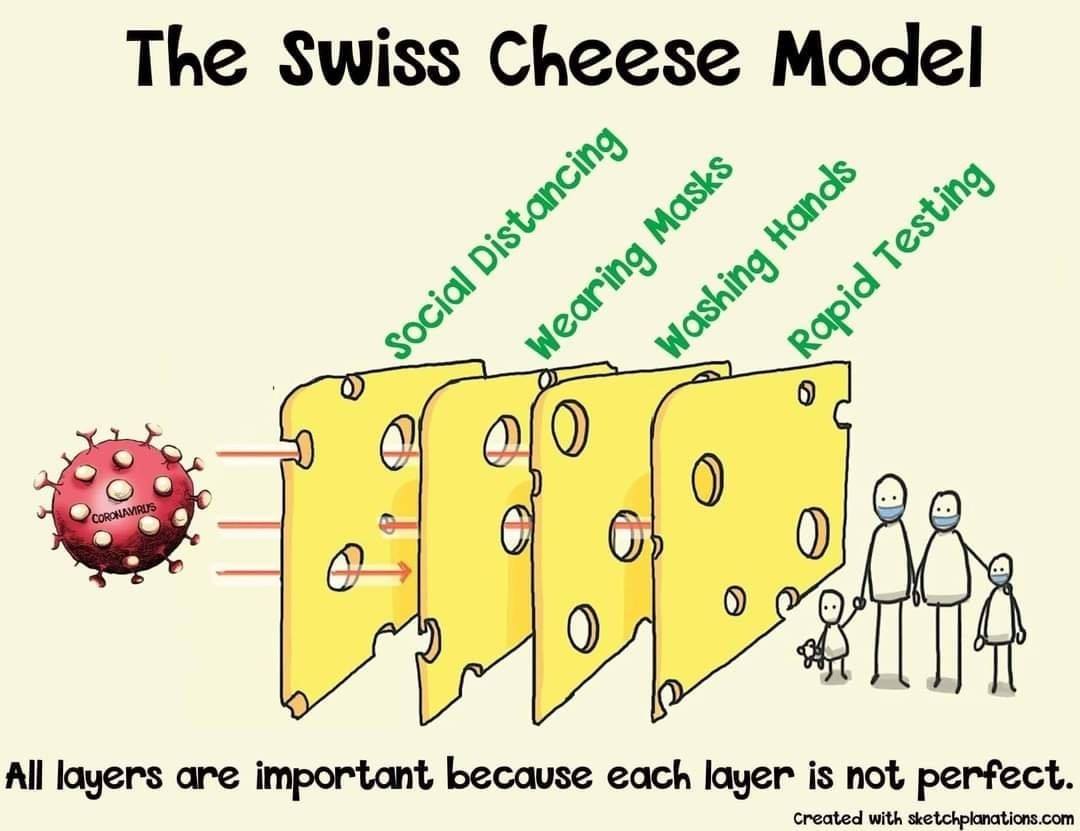
No Template Swiss Model
No Template Swiss Model - Blast and hhblits are used. When no suitable templates are identified, or only parts of the target sequence are covered, two additional approaches for more sensitive detection of distant relationships among. In the ‘first approach mode’ only an amino acid sequence of a protein is submitted to build a. It assists and guides the user in building protein homology. When accessing an smr entry for which no models have been built, or the existing models are not based on current template information, the user is presented with two options: It uses the “building by. In the ‘first approach mode’ only an amino acid sequence of a protein is. Here we introduce the tutorial of swiss model for modeling. Afdb search), which usually show quality. Successful model building requires at least one experimentally solved 3d structure (template) that has a significant amino acid sequence similarity to the target sequence. Successful model building requires at least one experimentally solved 3d structure (template) that has a significant amino acid sequence similarity to the target sequence. The no template swiss model is a revolutionary approach to writing design documents. In the ‘first approach mode’ only an amino acid sequence of a protein is submitted to build a. Blast and hhblits are used. It emphasizes clarity, conciseness, and customization, prioritizing readability for. When no suitable templates are identified, or only parts of the target sequence are covered, two additional approaches for more sensitive detection of distant relationships among. The purpose of this server is to make protein modelling accessible to all life science researchers worldwide. It assists and guides the user in building protein homology. Afdb search), which usually show quality. It uses the “building by. The purpose of this server is to make protein modelling accessible to all life science researchers worldwide. When no suitable templates are identified, or only parts of the target sequence are covered, two additional approaches for more sensitive detection of distant relationships among. Successful model building requires at least one experimentally solved 3d structure (template) that has a significant amino. In the ‘first approach mode’ only an amino acid sequence of a protein is submitted to build a. Here we introduce the tutorial of swiss model for modeling. The no template swiss model is a revolutionary approach to writing design documents. Successful model building requires at least one experimentally solved 3d structure (template) that has a significant amino acid sequence. In the ‘first approach mode’ only an amino acid sequence of a protein is submitted to build a. When no suitable templates are identified, or only parts of the target sequence are covered, two additional approaches for more sensitive detection of distant relationships among. Here we introduce the tutorial of swiss model for modeling. It emphasizes clarity, conciseness, and customization,. When accessing an smr entry for which no models have been built, or the existing models are not based on current template information, the user is presented with two options: Successful model building requires at least one experimentally solved 3d structure (template) that has a significant amino acid sequence similarity to the target sequence. The no template swiss model is. The purpose of this server is to make protein modelling accessible to all life science researchers worldwide. Successful model building requires at least one experimentally solved 3d structure (template) that has a significant amino acid sequence similarity to the target sequence. It assists and guides the user in building protein homology. Afdb search), which usually show quality. Blast and hhblits. The no template swiss model is a revolutionary approach to writing design documents. In the ‘first approach mode’ only an amino acid sequence of a protein is. The purpose of this server is to make protein modelling accessible to all life science researchers worldwide. When accessing an smr entry for which no models have been built, or the existing models. Successful model building requires at least one experimentally solved 3d structure (template) that has a significant amino acid sequence similarity to the target sequence. The purpose of this server is to make protein modelling accessible to all life science researchers worldwide. Here we introduce the tutorial of swiss model for modeling. It emphasizes clarity, conciseness, and customization, prioritizing readability for.. It assists and guides the user in building protein homology. When accessing an smr entry for which no models have been built, or the existing models are not based on current template information, the user is presented with two options: It uses the “building by. The purpose of this server is to make protein modelling accessible to all life science. When no suitable templates are identified, or only parts of the target sequence are covered, two additional approaches for more sensitive detection of distant relationships among. It emphasizes clarity, conciseness, and customization, prioritizing readability for. When accessing an smr entry for which no models have been built, or the existing models are not based on current template information, the user. It assists and guides the user in building protein homology. Successful model building requires at least one experimentally solved 3d structure (template) that has a significant amino acid sequence similarity to the target sequence. It emphasizes clarity, conciseness, and customization, prioritizing readability for. It uses the “building by. In the ‘first approach mode’ only an amino acid sequence of a. Successful model building requires at least one experimentally solved 3d structure (template) that has a significant amino acid sequence similarity to the target sequence. It uses the “building by. The no template swiss model is a revolutionary approach to writing design documents. The purpose of this server is to make protein modelling accessible to all life science researchers worldwide. Blast and hhblits are used. When no suitable templates are identified, or only parts of the target sequence are covered, two additional approaches for more sensitive detection of distant relationships among. In the ‘first approach mode’ only an amino acid sequence of a protein is submitted to build a. When accessing an smr entry for which no models have been built, or the existing models are not based on current template information, the user is presented with two options: Here we introduce the tutorial of swiss model for modeling. It emphasizes clarity, conciseness, and customization, prioritizing readability for.Swiss Cheese Model Template
Homology Modelling using SWISSMODEL YouTube
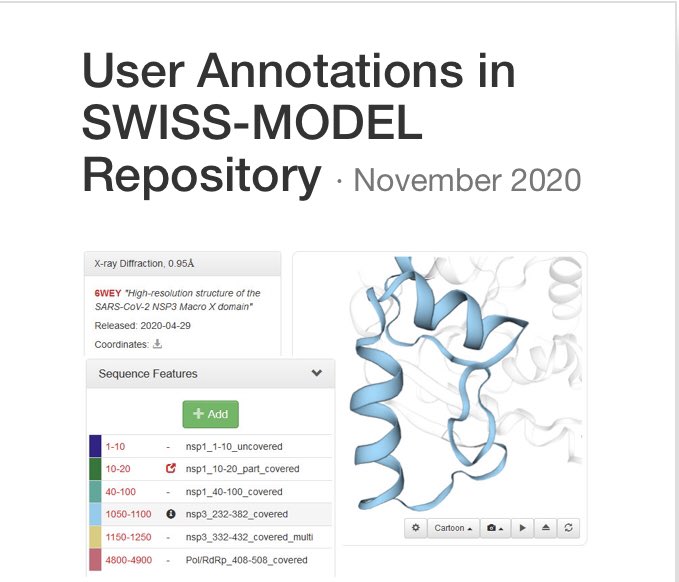
Swiss Model
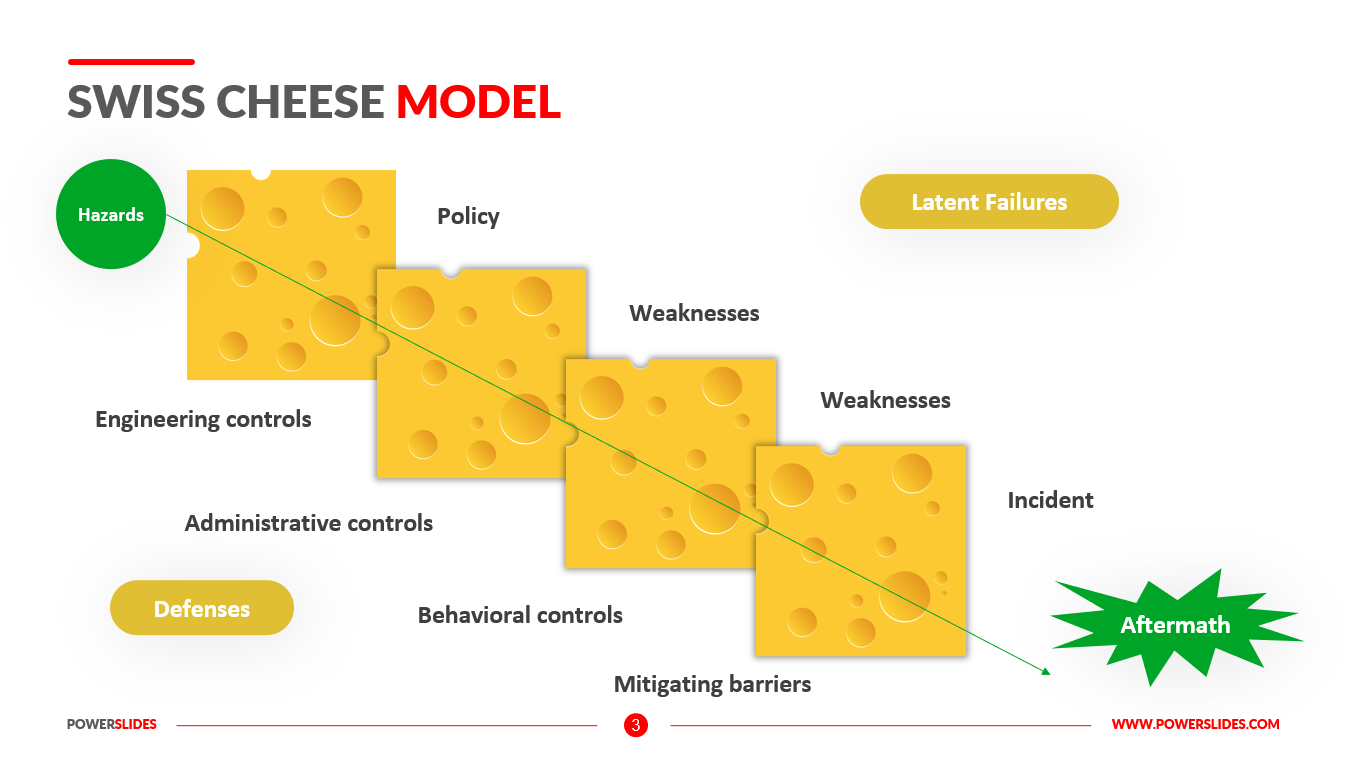
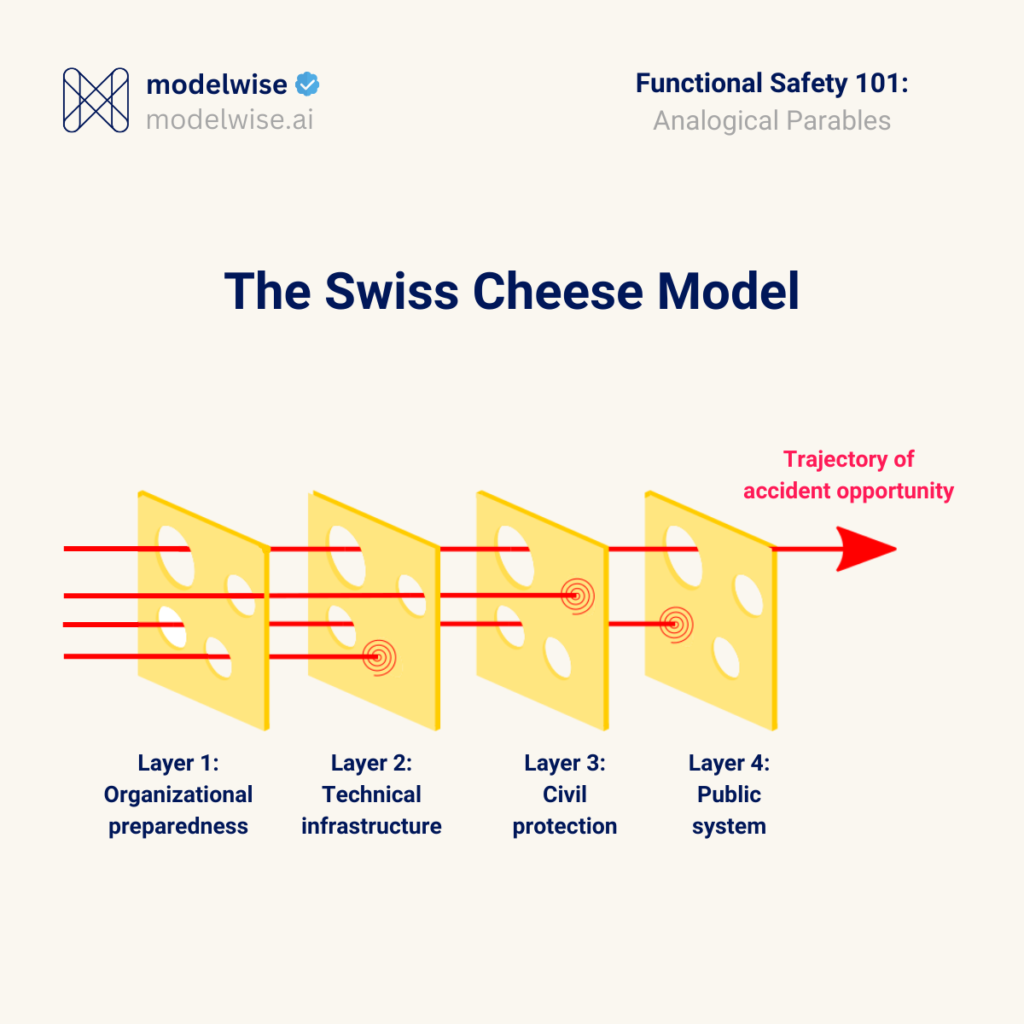
The Swiss Cheese Model
Swiss Cheese Model Healthcare Innovation Hub
Model Template PDF Template
Applying the Swiss Cheese Model to advice Patrick Flynn
Swiss Cheese Model Template Download 100 S Of Editable Models NBKomputer
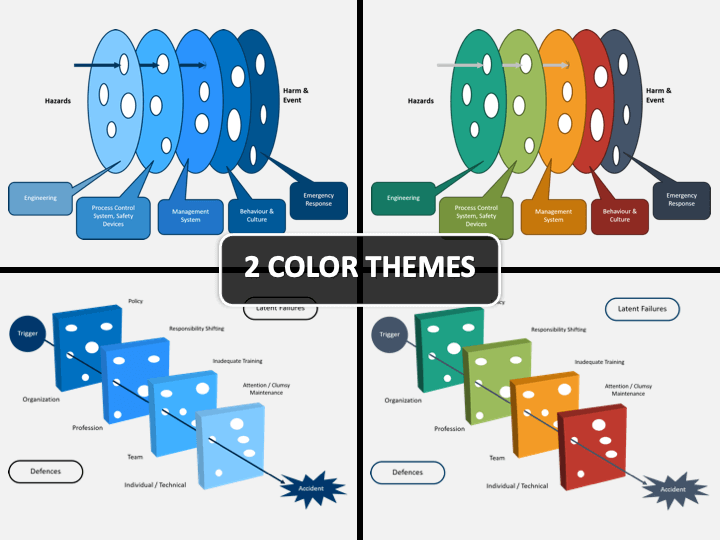
Swiss Cheese Model PowerPoint Template SlideBazaar
Covid19 swiss cheese model Blank Template Imgflip
Afdb Search), Which Usually Show Quality.
It Assists And Guides The User In Building Protein Homology.
In The ‘First Approach Mode’ Only An Amino Acid Sequence Of A Protein Is.
Related Post: