No Template Alphafold
No Template Alphafold - You can try to input the sequence of a new protein target and see predict it using alphafold, by performing some changes to the input like template selection, or obtain a more diverse. Cosmic² offers the full alphafold2 software package for use by the structural. Is it possible to use an alphafold predicted structure as templates for picking? The template acts as a reference, nudging. It uses a novel machine learning approach to predict 3d protein structures from primary. Sequence alignments/templates are generated through mmseqs2 and hhsearch. Secondly, i did an ugly hack on the alphafold/data/msa_pairing.py file in order to run the multimer pipeline without templates. If you want to run the pipeline with custom msa/templates, you need to set all of them. Sequence alignments/templates are generated through mmseqs2 and hhsearch. Alphafold is a protein structure prediction tool developed by deepmind (google). When this option is true, colabfold will search the pdb sequences for similarity to the target and report in the colab log which entries. If you want to run the pipeline with custom msa/templates, you need to set all of them. Using the none option will result in no template being used, the pdb70 option results in a similar structure. It uses a novel machine learning approach to predict 3d protein structures from primary. Sequence alignments/templates are generated through mmseqs2 and hhsearch. Alphafold2 leverages multiple sequence alignments and neural networks to predict protein structures. Alphafold2 uses templates as a sort of guide to help it improve its prediction. Sequence alignments/templates are generated through mmseqs2 and hhsearch. Alphafold is a protein structure prediction tool developed by deepmind (google). In our recent article applying and improving alphafold at casp14 (preview) we describe for the casp14 target t1024 that we had the same issue where alphafold ignored templates with. Secondly, i did an ugly hack on the alphafold/data/msa_pairing.py file in order to run the multimer pipeline without templates. Sequence alignments/templates are generated through mmseqs2 and hhsearch. You can try to input the sequence of a new protein target and see predict it using alphafold, by performing some changes to the input like template selection, or obtain a more diverse.. Alphafold can use up to four structures as templates; As of this latest release, pdb structures shown to the model are recorded in. Using the none option will result in no template being used, the pdb70 option results in a similar structure. Alphafold2 uses templates as a sort of guide to help it improve its prediction. Alphafold2 leverages multiple sequence. Alphafold is a protein structure prediction tool developed by deepmind (google). You can provide structure models (preferably in the mmcif format) as templates to guide alphafold2 to predict a protein in a specific state. Alphafold2 uses templates as a sort of guide to help it improve its prediction. Alphafold can use up to four structures as templates; Is it possible. If you want to run the pipeline with custom msa/templates, you need to set all of them. I am trying to run alphafold using a custom template. Sequence alignments/templates are generated through mmseqs2 and hhsearch. You can set msa to empty string and templates to empty list to signify that they should. In our recent article applying and improving alphafold. Model 1.1.1 of alphafold (default setting) 1 was used for the predictions, with no structural templates. What i did was basically tracking how the. If you want to run the pipeline with custom msa/templates, you need to set all of them. Alphafold is a protein structure prediction tool developed by deepmind (google). Cosmic² offers the full alphafold2 software package for. Secondly, i did an ugly hack on the alphafold/data/msa_pairing.py file in order to run the multimer pipeline without templates. Sequence alignments/templates are generated through mmseqs2 and hhsearch. Using the none option will result in no template being used, the pdb70 option results in a similar structure. Sequence alignments/templates are generated through mmseqs2 and hhsearch. You can try to input the. You can try to input the sequence of a new protein target and see predict it using alphafold, by performing some changes to the input like template selection, or obtain a more diverse. Secondly, i did an ugly hack on the alphafold/data/msa_pairing.py file in order to run the multimer pipeline without templates. You can set msa to empty string and. Alphafold can use up to four structures as templates; You can try to input the sequence of a new protein target and see predict it using alphafold, by performing some changes to the input like template selection, or obtain a more diverse. Alphafold is a protein structure prediction tool developed by deepmind (google). Cosmic² offers the full alphafold2 software package. Alphafold2 leverages multiple sequence alignments and neural networks to predict protein structures. When this option is true, colabfold will search the pdb sequences for similarity to the target and report in the colab log which entries. I am trying to run alphafold using a custom template. Hi dario, you could use the molmap feature in chimerax to generate a volume. It uses a novel machine learning approach to predict 3d protein structures from primary. In our recent article applying and improving alphafold at casp14 (preview) we describe for the casp14 target t1024 that we had the same issue where alphafold ignored templates with. What i did was basically tracking how the. Cosmic² offers the full alphafold2 software package for use. Alphafold can use up to four structures as templates; In our recent article applying and improving alphafold at casp14 (preview) we describe for the casp14 target t1024 that we had the same issue where alphafold ignored templates with. Model 1.1.1 of alphafold (default setting) 1 was used for the predictions, with no structural templates. It uses a novel machine learning approach to predict 3d protein structures from primary. Using the none option will result in no template being used, the pdb70 option results in a similar structure. Cosmic² offers the full alphafold2 software package for use by the structural. Sequence alignments/templates are generated through mmseqs2 and hhsearch. Alphafold is a protein structure prediction tool developed by deepmind (google). What i did was basically tracking how the. You can provide structure models (preferably in the mmcif format) as templates to guide alphafold2 to predict a protein in a specific state. Sequence alignments/templates are generated through mmseqs2 and hhsearch. As of this latest release, pdb structures shown to the model are recorded in. Secondly, i did an ugly hack on the alphafold/data/msa_pairing.py file in order to run the multimer pipeline without templates. Alphafold2 uses templates as a sort of guide to help it improve its prediction. You can try to input the sequence of a new protein target and see predict it using alphafold, by performing some changes to the input like template selection, or obtain a more diverse. The template acts as a reference, nudging.AlphaFold an AI tool for protein structure prediction Genenames Blog
AlphaFold performance at a range of template sequence... Download
Benchmarking AlphaFold for protein complex modeling reveals accuracy
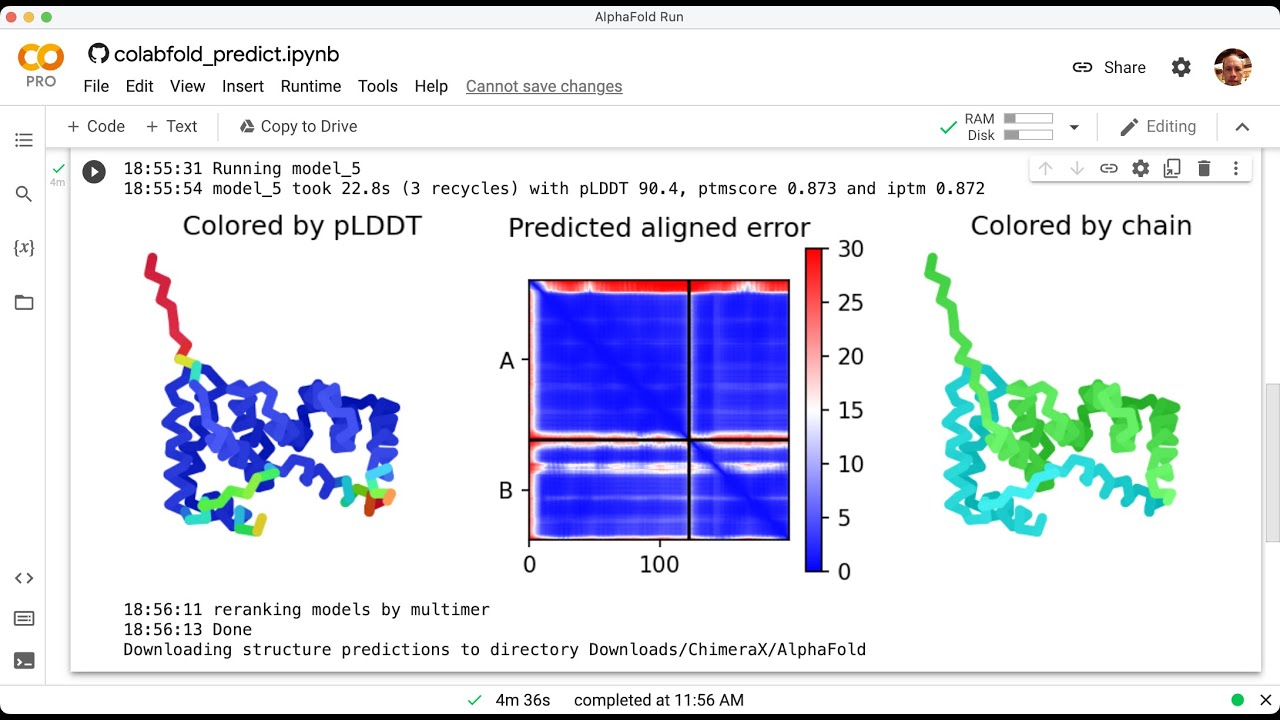
Faster AlphaFold protein structure predictions using ColabFold YouTube
What Is AlphaFold? New England Journal of Medicine
The Ultimate StepbyStep Guide to Using AlphaFold for Antibody Design
AlphaFold performance at a range of template sequence... Download
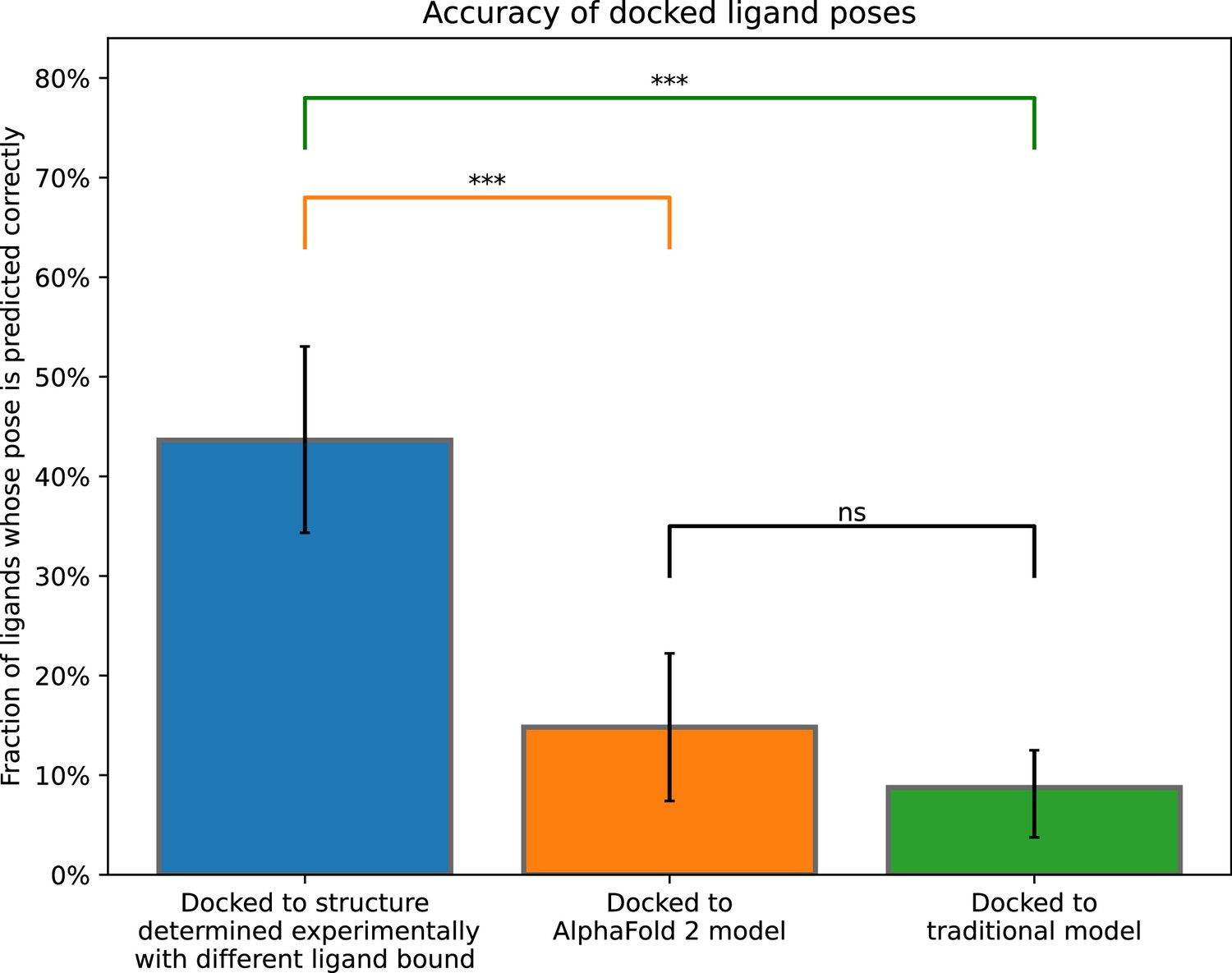
How accurately can one predict drug binding modes using AlphaFold
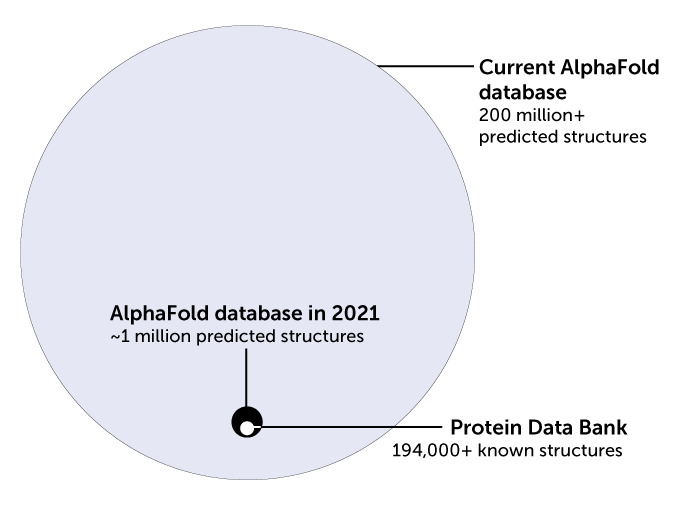
AlphaFoldbased databases and fullyfledged, easytouse AlphaFold
¿Ha resuelto AlphaFold el problema del plegamiento de proteínas de la
Sequence Alignments/Templates Are Generated Through Mmseqs2 And Hhsearch.
Is It Possible To Use An Alphafold Predicted Structure As Templates For Picking?
Alphafold2 Leverages Multiple Sequence Alignments And Neural Networks To Predict Protein Structures.
When This Option Is True, Colabfold Will Search The Pdb Sequences For Similarity To The Target And Report In The Colab Log Which Entries.
Related Post: